You are here
Omics
Omic technologies provide unique opportunities for the discovery of a new generation of biomarkers of exposure and disease risk, significantly enriched by mechanistic information, by permitting the simultaneous analysis of large numbers of potential targets without recourse to prior hypotheses. EXPOsOMICS advances omic technologies that are related to identification and quantification of environmental exposure (metabolomics, adductomics) and applies multiplex omic technologies that measure downstream effects of environmental exposures (epigenome, transcripts, miRNA, proteins).
Omic technologies are deployed in an untargeted manner in both the short-term and the long-term studies (Phase 1), to a total of approximately 3,000 samples.
The subjects for untargeted omics in Phase 1 were selected on the basis of extreme exposures. Publicly available toxicogenomics databases have been explored to identify candidate omic-based exposure biomarkers. Databases are the diXA data infrastructure (FP7 project: diXA) and the US Comparative Toxicogenomics Data Base (http://ctdbase.org/). These contain both in vitro and in vivo toxicogenomic data and, to a degree, omic results from exposed humans, and have links to important chemical toxicity databases such as the US NTP and the OECD eChemPortal. We will attempt to replicate the signals of phase 1 in selected case-cohort studies for a total of approximately 2,000 subjects. One of the main products of the consortium will be endophenotypes, i.e. intermediate phenotypes common to several diseases.
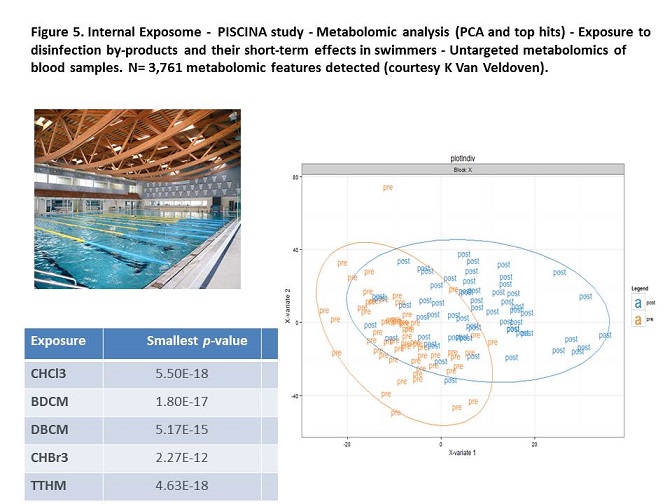
More details>>

